Climate change is likely to have profound impacts on ecological systems, but in some important cases we’re still a long way from understanding exactly what those impacts might be. For instance, it has been posited that under warming, species in the northern hemisphere will be enhanced at their northern range edge (where presumably species are fitness-limited, and hence range-limited, by the cooler temperatures). Rather, recent research indicates that individual populations of some species may be locally adapted to their climatic environment, such that even these peripheral populations may decline with warming as they have developed an equilibrium with their environment.
Here, we examine gene-expression profiles of these populations for two butterfly species. Even though both species were highly differentiated between populations, for the species that shows more population structure as well as more evidence for local adaptation we discovered a variety of genes responding uniquely to climate differences. Functions of these genes ranged from energy metabolism to detoxification of food sources, suggesting that the effects of climate change may be strongly linked to species’ locations and roles in their ecological networks. (Paper.)
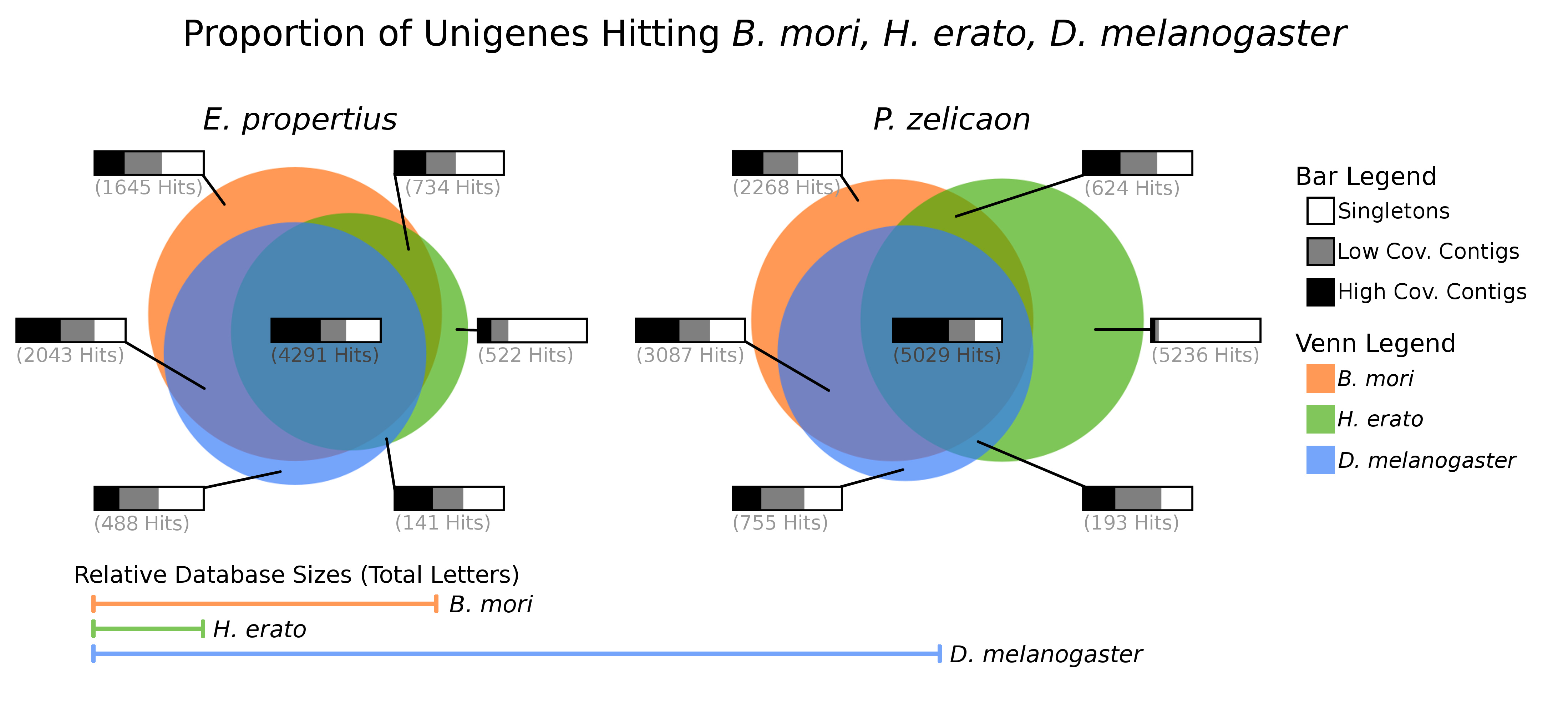
In transcriptome sequencing and de-novo assembly, we sequence messenger RNA in short fragments and reassemble them into longer sequences representing coding genes. Using sequencing similarity tools, we can then compare the gene sets to gene sets of related organisms. For the project above, we produced transcriptomes for the two butterfly species, and compared them to gene sets for the fruit fly (D. melanogaster) silk worm (B. mori) and another butterfly (H. erato). Interestingly, we found that a large number of unassembled Swallowtail fragments matched only H. erato (large green area, right); these turned out to be ribosomal RNA (which is not polyadenylated and hence contamination). (Paper.)